The world is facing a major sixth wave of extinction, experts say. The five previous waves occurred already millions of years ago: the first extinction wave about 443 million years ago, with a large sea level drop as a result of cooling (Appalachian formation probably played a role in this). This is also called the Ordovician/Silurian extinction. Many marine organisms such as corals and trilobites became extinct in this wave. The second occurred at the end of the Devonian era, about 359 million years ago. The cause was probably a large decrease in the oxygen content in the oceans, possibly caused by volcanism. Many marine organisms, such as precursors of current mollusks, also died in this wave. The third wave, which marked the transition from the Permian to Triassic era, meant the extinction of many animal species. This has been the largest extinction wave to date (almost all marine life and three quarters of life on land died out). This wave, some 251 million years ago, according to researchers is due to a large-scale release of carbon dioxide, much more than we currently release into the atmosphere. The fourth wave marked the transition from the Triassic to Jurassic era - some 201 million years ago. Here too, the release of a mega amount of CO2 plays a major role (as a result of magma released on the earth's surface). This extinction wave meant the beginning of the dinosaur era, which, with the impact of an asteroid, came to a sudden end some 66 million years ago and made way for the mammals. This transition, from Cretaceous to Paleogene (known as the K-Pg transition) is called the fifth great extinction and is most well-known to the general public. For more detailed information on extinction waves, see Greshko's article in National Geographic (2019), or the information on the website of the American Museum of Natural History. For a clear scientific article: Barnosky, Nature 2011.
An article will be published in iScience on May 20 in which a relatively recent methodology is used to get a better grip on biodiversity. By learning more about numbers of species in the past and present, one can draw stronger conclusions about the sixth extinction wave and the relationship with human activities. The method is called palaeoproteomics and focuses on the proteins in ancient to very ancient biological material. It is often very difficult to identify the animal or plant species on the basis of fossil remains. The materials are often damaged, not intact or largely petrified. And, isolating DNA is often no longer possible, because DNA breaks down and can no longer be analyzed after about 1.5 million years. In the article by Peters and colleagues, the method is called 'conservation palaeoproteomics' and possibilities are explored using this method to get a better grip on, for example, invasive species, illegal trade in flora or fauna, biogeography of species and the influence of humans on the environment. present flora and fauna. The authors expect that the method will take center stage in many areas. Consider, for example, archaeology, where animal materials in paint or milk residues can be analyzed. At Naturalis in Leiden, we have also worked with palaeoproteomics to study the extinct dodo and the great auk – a bird that was once numerous in the Northern Hemisphere coastal regions and present on the Dutch North Sea coast as well. We were able to identify the species and determine relationships with other bird species.
Proteins are thus more resistant to the test of time. This is not so much because these molecules are stronger, but because of the environment in which they find themselves when an organism fossilizes. Think of bone or tooth material, where the structure provides protection to these molecules. And they are so for a very long time: there are studies out there that looked at 195 million year old proteins. The question remains whether the materials are not contaminated with protein of a more recent date. In any case, we already know that we can see much further than the 1.5 million years that DNA allows us!
How do we do that? The first "palaeobiochemical" studies were described in 1957 by the American physicist Philip Abelson (discoverer of one of the elements and participant in the Manhattan project). At that time, amino acids, the building blocks of proteins, were separated by means of paper chromatography. Abelson described the presence of, among other things, amino acids in fossil remains. Due to the absorbing effect of paper, molecules on paper filters can be separated by size (when using the right solutions). Today, analysis is very different: we measure all atoms (actually all atomic masses) in the molecules we have in a preparation and compare the data with a very large database. The method most commonly used here is mass spectrometry. In palaeoproteomics – all proteins present ('the proteome') in very old material ('palaeo') - we use a mass spectrometer to measure which atoms are present. And by precisely determining the masses. So we know exactly which molecule we have in a preparation after database identification (see also figure 1 from the article by Peters). And because databases in biology grow exponentially - the field of 'big data' - we can also compare more and more. However, this method is not the only one; researchers also use complex spectroscopy with usually unpronounceable long names. For example, in situ Synchotron FTIR microspectroscopy. This is a method in which you analyze a sample on the spot (in situ, without first isolating molecules in a lab), using infrared light. The synchotron addition (a particle accelerator) ensures that the spectroscopy produces even better pictures with higher resolution. You actually scan a sample with this equipment and you immediately determine which molecules are present. Chemically speaking, the bonds between atoms are investigated: when irradiated with infrared, they start to vibrate in a bond-specific way. The method is also widely used in biomedical research to distinguish cancer cells from normal cells, for example. Finally, antibodies are in use as well. These are our ‘defense proteins’ that are widely used in biological and biomedical research. For example, there are antibodies that specifically bind to proteins in bone tissue. By attaching a dye (a color) to the antibody, you can visualize the protein in a sample.
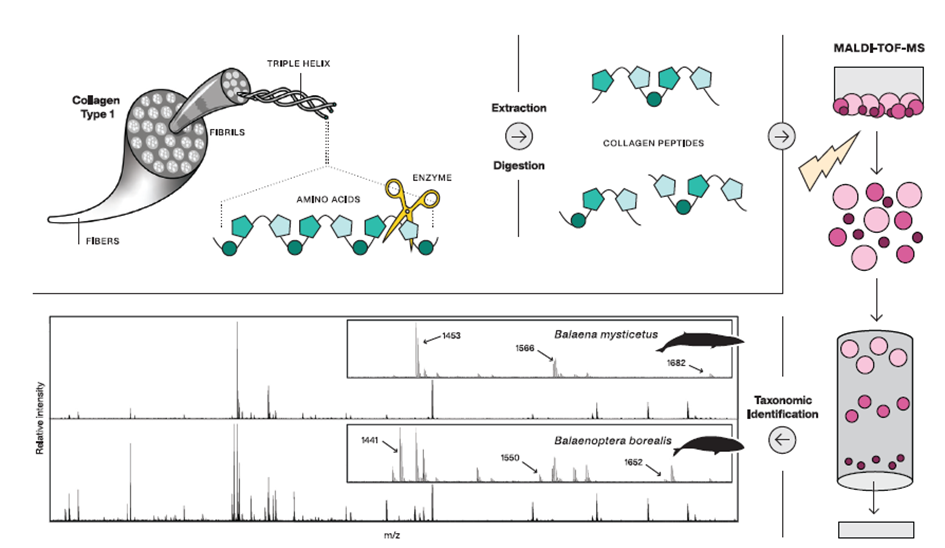
Figure 1. Palaeoproteomics analysis of proteins in a sample. The proteins are first cut with an enzyme, then placed in a mass spectrometer (in this case a 'MALDI-TOF' instrument), analyzed and the spectrum is analyzed. This ultimately leads to identification of the species. (Figure from Peters, 2022, with permission).
More information:
- Peters, 2022, https://doi.org/10.1016/j.isci.2022.104195
A.D. Barnosky, 2011, https://doi.org/10.1038/nature09678
https://www.nationalgeographic.com/science/article/mass-extinction
https://www.amnh.org/exhibitions/dinosaurs-ancient-fossils/extinction/mass-extinction
I.R. Horn, 2019, https://doi.org/10.1093/zoolinnean/zlz012
P.H. Abelson, 1955, Some aspects of paleobiochemistry. Annals of the New York Academy of Sciences.
Y-C. Lee, 2017, http://dx.doi.org/10.1038/ncomms14220


Add comment
Comments